www.georginajoyce.com
And thanks to everyone at @cmrqut.bsky.social for your support and @emerge-bii.bsky.social for funding this research!

www.georginajoyce.com
And thanks to everyone at @cmrqut.bsky.social for your support and @emerge-bii.bsky.social for funding this research!
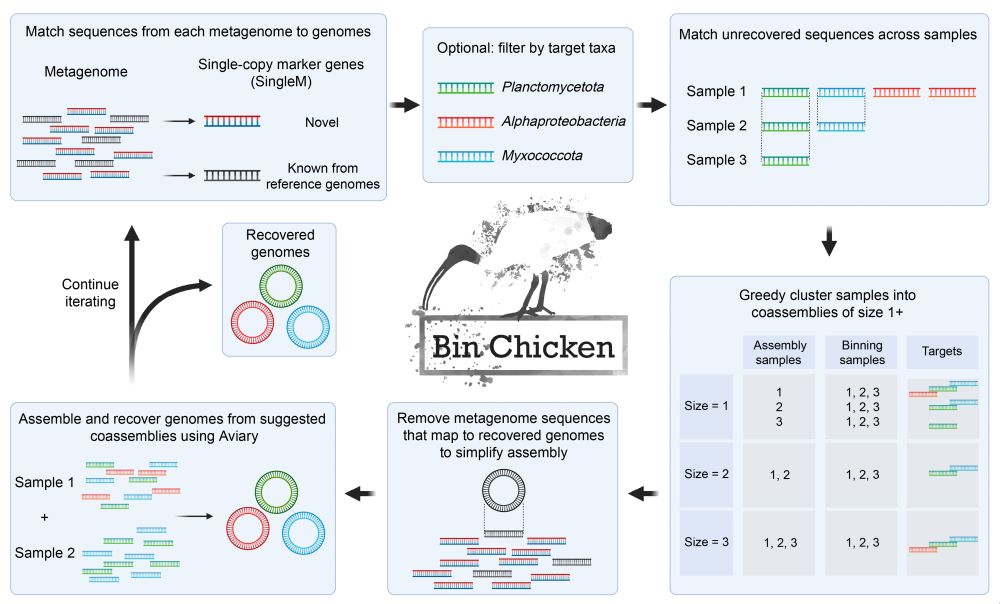
Visit github.com/aroneys/binchicken to try it for yourself! 5/

Visit github.com/aroneys/binchicken to try it for yourself! 5/


🌐 uhgv.jgi.doe.gov (8/8)
🌐 uhgv.jgi.doe.gov (8/8)

