www.nature.com/articles/s41...
Caviar for RNA-Seq nerds! Check this out

www.nature.com/articles/s41...
Caviar for RNA-Seq nerds! Check this out

Something different from the group! Thanks to Yuxin, a talented student in the lab, we used long-reads in 75 human trios to study telomeres and their inheritance.

Something different from the group! Thanks to Yuxin, a talented student in the lab, we used long-reads in 75 human trios to study telomeres and their inheritance.
Special thanks to the reviewers whose comments improved our manuscript a lot! rdcu.be/eI3tD

Special thanks to the reviewers whose comments improved our manuscript a lot! rdcu.be/eI3tD
Delighted to share the story of two germline RBPs - one with little (DND1) and one with no (NANOS3) intrinsic sequence-specificity - that together build a continuous RNA binding surface recognizing a 7-mer (AUGAAUU) in target mRNA 3’UTRs, leading to deadenylation.

Delighted to share the story of two germline RBPs - one with little (DND1) and one with no (NANOS3) intrinsic sequence-specificity - that together build a continuous RNA binding surface recognizing a 7-mer (AUGAAUU) in target mRNA 3’UTRs, leading to deadenylation.

www.nature.com/articles/s41...
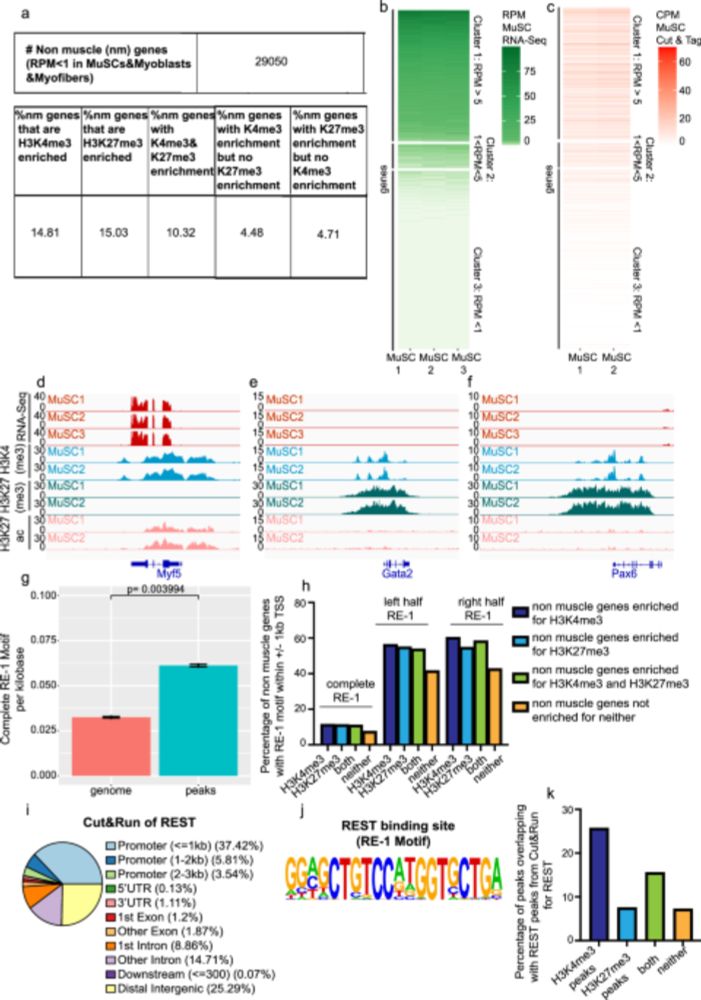
www.nature.com/articles/s41...
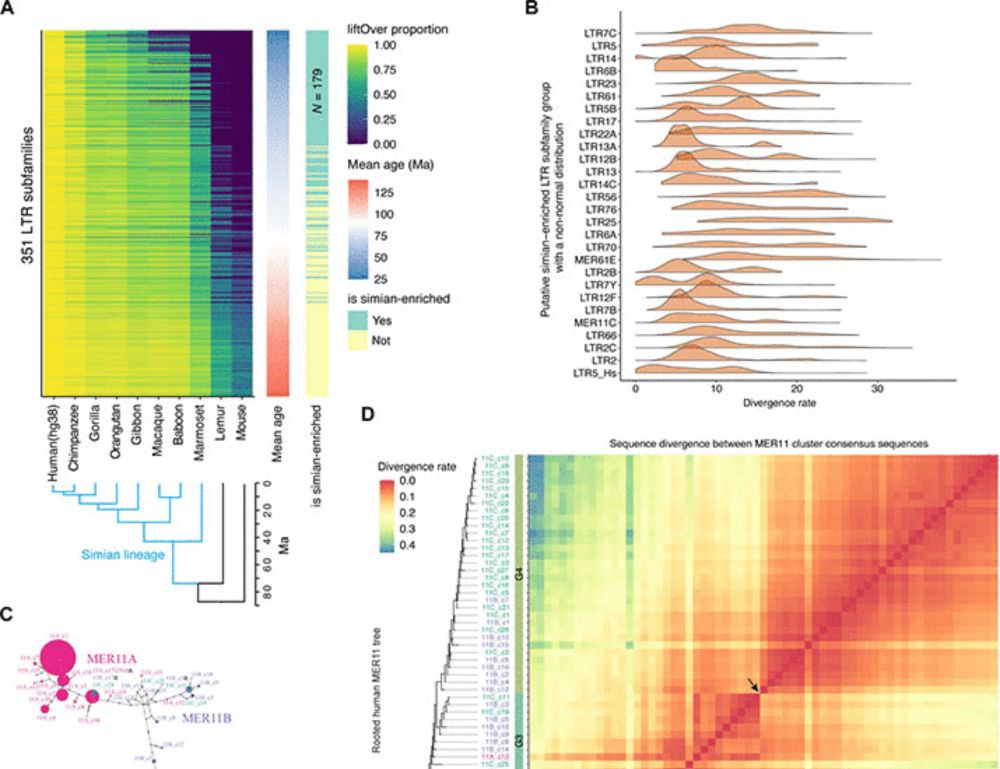
@natbiotech.nature.com: nature.com/articles/s41.... A short tread! (1/n)
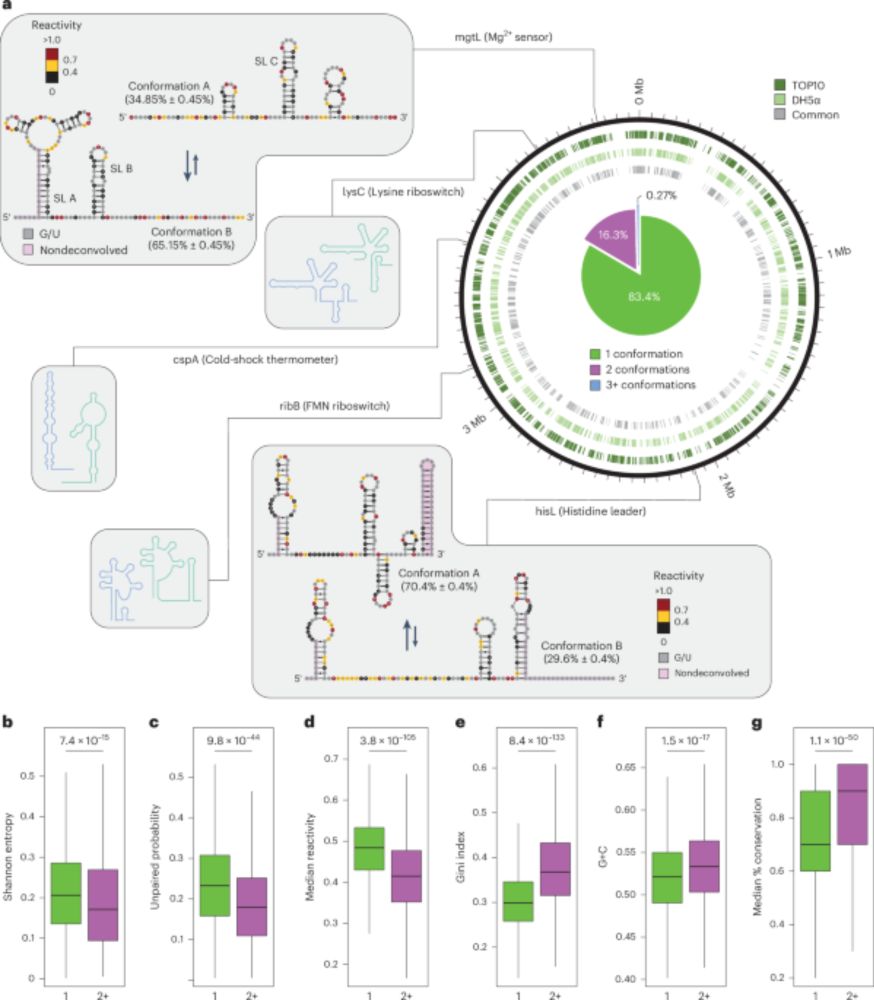
@natbiotech.nature.com: nature.com/articles/s41.... A short tread! (1/n)
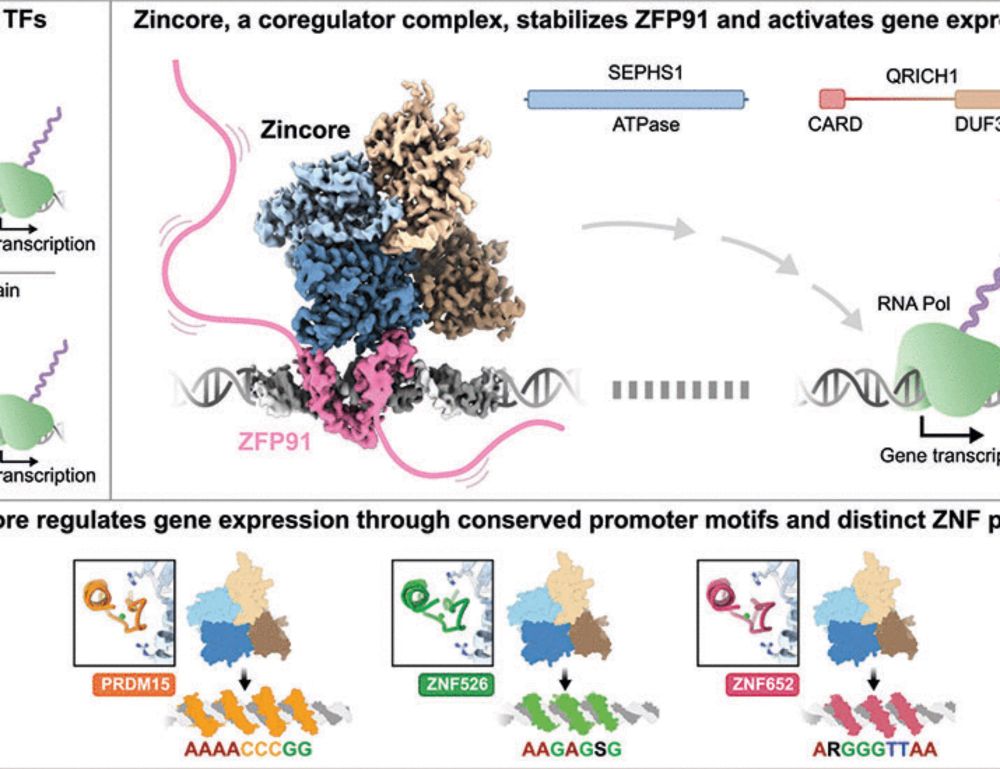
July 10th, 1pm-4pm ET ONLINE for everyone
July 11th, 9am-7pm ET ONLINE across Canada and IN PERSON in Toronto
event.fourwaves.com/trend25/regi...

🔗 doi.org/10.1093/bioa...
#Genomics #Bioinformatics #RegulatoryGenomics

🔗 doi.org/10.1093/bioa...
#Genomics #Bioinformatics #RegulatoryGenomics


What determines which AS isoforms result in abundant proteins ?
We find that proteoform-specific ...
🧵
www.biorxiv.org/content/10.1...
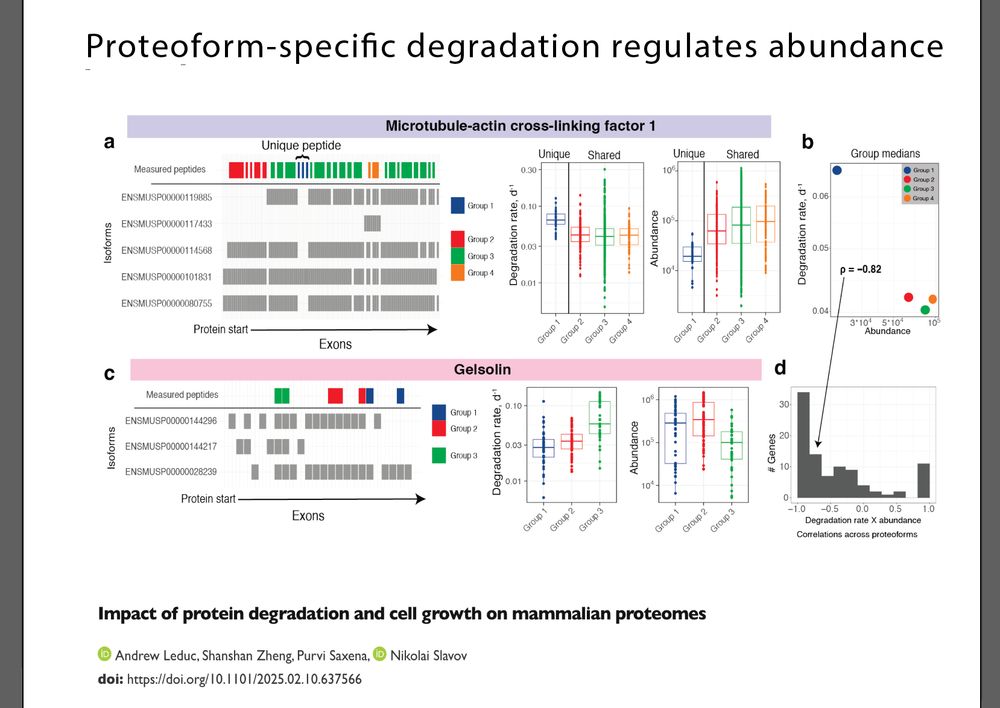
What determines which AS isoforms result in abundant proteins ?
We find that proteoform-specific ...
🧵
www.biorxiv.org/content/10.1...
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
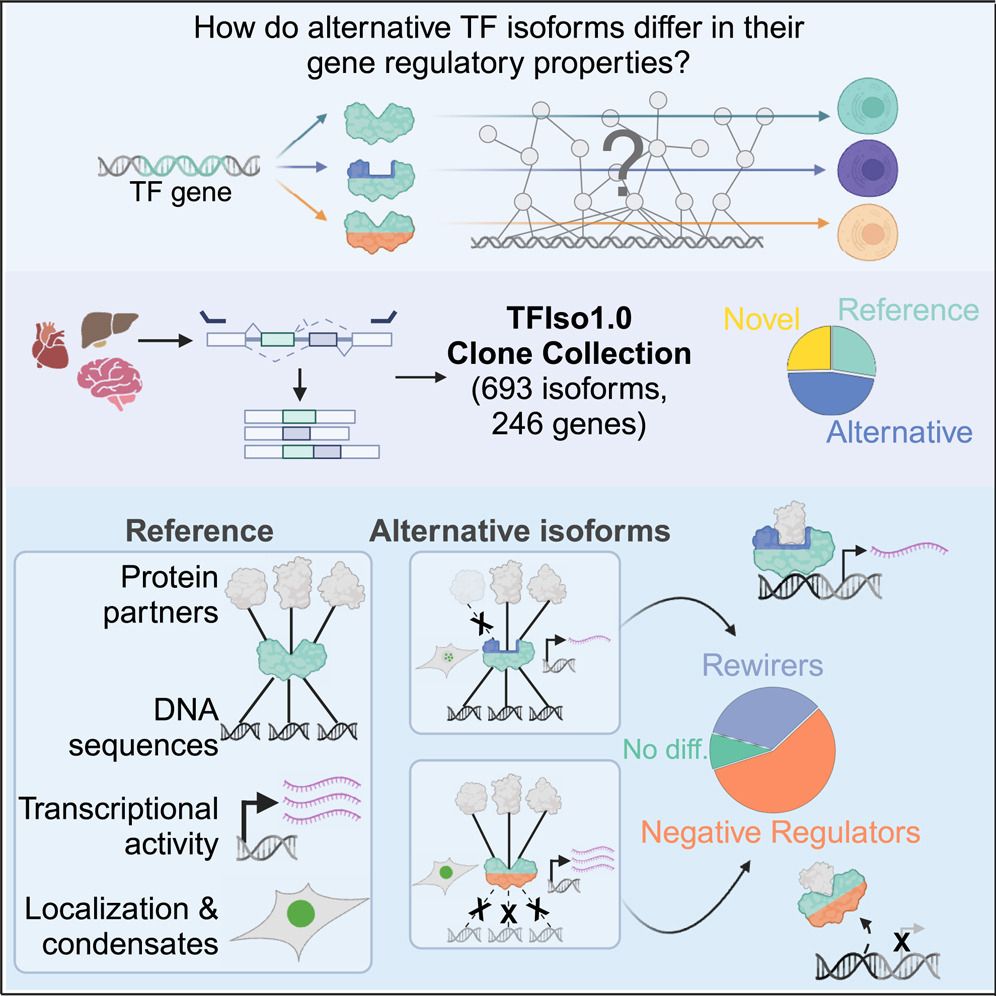
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

Our latest preprint, led by Junbum Kim, maps early-stage cancer at single-cell resolution, revealing two key pathways:
🦠Driven by immune activity
🛠️ Driven by fibrosis
These insights could refine detection & treatment.
www.biorxiv.org/content/10.1...

Our latest preprint, led by Junbum Kim, maps early-stage cancer at single-cell resolution, revealing two key pathways:
🦠Driven by immune activity
🛠️ Driven by fibrosis
These insights could refine detection & treatment.
www.biorxiv.org/content/10.1...
Check out our paper here: www.biorxiv.org/content/10.1...
Check out our paper here: www.biorxiv.org/content/10.1...
Catch the full conversation:
+ YouTube: youtu.be/sWcoslBueiE
+ Spotify: spoti.fi/4hLk3xo
+ Apple Podcasts: apple.co/40sQCu9
Catch the full conversation:
+ YouTube: youtu.be/sWcoslBueiE
+ Spotify: spoti.fi/4hLk3xo
+ Apple Podcasts: apple.co/40sQCu9
By investigating bacterial genomes put in yeast, we show that the presence or absence of transcription is sufficient!
#chromatin #3Dgenome #generegulation
www.science.org/doi/10.1126/...
👇
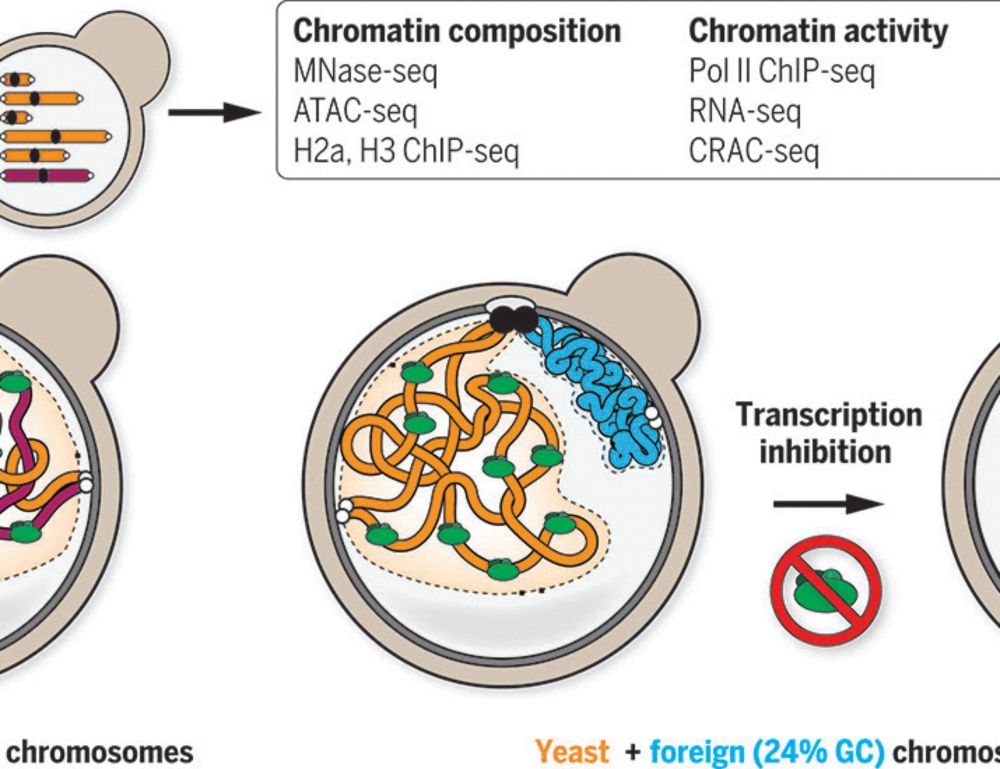
By investigating bacterial genomes put in yeast, we show that the presence or absence of transcription is sufficient!
#chromatin #3Dgenome #generegulation
www.science.org/doi/10.1126/...
👇

www.encodeproject.org/single-cell/...
www.encodeproject.org/single-cell/...


