
Congrats @erezyirmiya.bsky.social and @soreklab.bsky.social
www.biorxiv.org/content/10.6...

Congrats @erezyirmiya.bsky.social and @soreklab.bsky.social
www.biorxiv.org/content/10.6...

It was a great honor to contribute to this project! Congratulations to @erezyirmiya.bsky.social and Azita Leavitt!!🎉🥂
www.biorxiv.org/content/10.6...

It was a great honor to contribute to this project! Congratulations to @erezyirmiya.bsky.social and Azita Leavitt!!🎉🥂
Conservation of TIR-derived signals accross the tree of life! We found bacterial TIR immune systems that signal via canonical cADPR (like in humans) and 2'cADPR (a plant immune signal).
Documented 11 Thoeris types

Conservation of TIR-derived signals accross the tree of life! We found bacterial TIR immune systems that signal via canonical cADPR (like in humans) and 2'cADPR (a plant immune signal).
Documented 11 Thoeris types
Congrats @erezyirmiya.bsky.social & Azita! 👏🌟
www.biorxiv.org/content/10.6...

Congrats @erezyirmiya.bsky.social & Azita! 👏🌟
www.biorxiv.org/content/10.6...

www.biorxiv.org/content/10.6...

www.biorxiv.org/content/10.6...
Here’s the story of how we discovered the Metis defense system 👇
www.biorxiv.org/content/10.1...

We designed synthetic proteins that can block bacterial immune systems, allowing phages + plasmids to overcome natural defenses.
This could transform phage therapy + genetic engineering.
Here’s what we found 🧵
Preprint🔗: www.biorxiv.org/content/10.1...

We designed synthetic proteins that can block bacterial immune systems, allowing phages + plasmids to overcome natural defenses.
This could transform phage therapy + genetic engineering.
Here’s what we found 🧵
Preprint🔗: www.biorxiv.org/content/10.1...
Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵

Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵
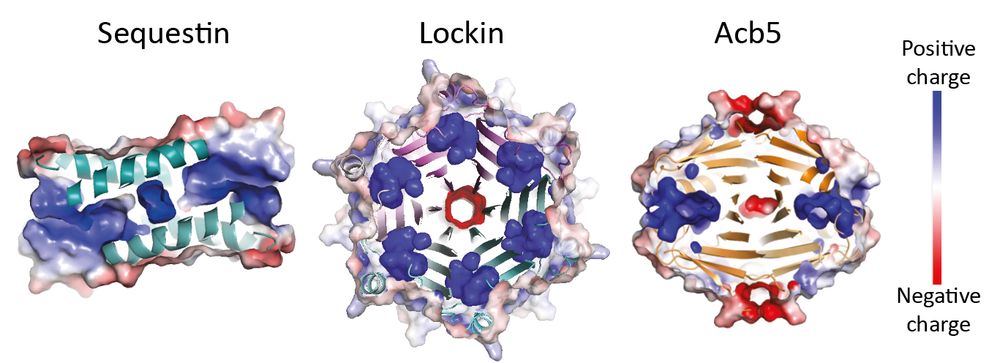
Together with @reneechang.bsky.social @kranzuschlab.bsky.social and the amazing @soreklab.bsky.social, we explored viral sponges to map their diversity and function.
Discovered huge diversity, including sponges that inhibit Pycsar & Type IV Thoeris!
www.biorxiv.org/content/10.1...

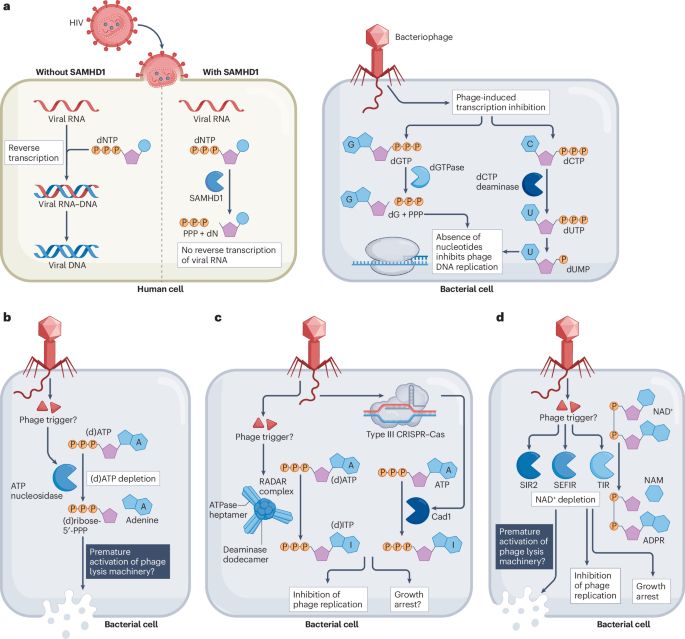
& how viruses overcome these nucleotide-based immune mechanisms 🗡️
www.nature.com/articles/s41...
Congrats Nitzan and everyone!
Excited to share my final work from the @soreklab.bsky.social!
We mined phage dark matter using structural features shared by anti-defense proteins (viral tools that help phages bypass bacterial immunity) to guide discovery.
Found 3 new families targeting immune signaling!
Congrats Nitzan and everyone!

search.app/jDxaUi5yJK9B...

search.app/jDxaUi5yJK9B...
Bacteria have been amazing ressources of molecular diversity for biotech and medicine, now viral diversity can be explored too!
Bacteria have been amazing ressources of molecular diversity for biotech and medicine, now viral diversity can be explored too!
Remarkable that phage proteins can inhibit human and plant TIR proteins and human cGAS. Conservation of function across such long evolutionary distances identifies new immunomodulators
Congrats to @soreklab.bsky.social and Kranzusch labs! What a great collaboration that is 😍
🧪
Remarkable that phage proteins can inhibit human and plant TIR proteins and human cGAS. Conservation of function across such long evolutionary distances identifies new immunomodulators
Congrats to @soreklab.bsky.social and Kranzusch labs! What a great collaboration that is 😍
🧪
Big congrats @erezyirmiya.bsky.social et al!!
Big congrats @erezyirmiya.bsky.social et al!!
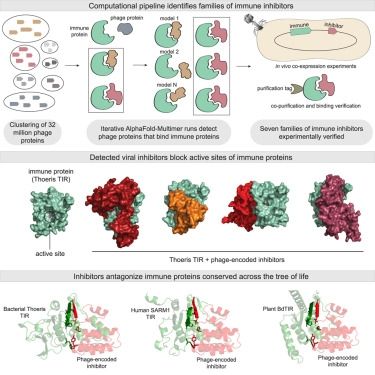
@erezyirmiya et al identified viral proteins that inhibit host immunity using structural predictions of protein-protein interactions.
Congrats to you all @soreklab & Kranzusch lab! 🥂
@erezyirmiya et al identified viral proteins that inhibit host immunity using structural predictions of protein-protein interactions.
Congrats to you all @soreklab & Kranzusch lab! 🥂
Congrats @erezyirmiya.bsky.social @soreklab.bsky.social et al for these beautiful discoveries 👌
Congrats @erezyirmiya.bsky.social @soreklab.bsky.social et al for these beautiful discoveries 👌
Congrats Erez Yirmiya, Azita Leavitt, Gil Amitai, our collaborators at the Kranzusch lab, and coauthors
A 🧵 1/10
www.cell.com/cell/abstrac...
Congrats Erez Yirmiya, Azita Leavitt, Gil Amitai, our collaborators at the Kranzusch lab, and coauthors
A 🧵 1/10
www.cell.com/cell/abstrac...

