Yousuf A. Khan
@yousufakhan.bsky.social
1.4K followers
250 following
51 posts
Group Leader @Stanford. RNA focused ML/AI, cryoEM/ET, and biophysics. Formerly:DeepMind AlphaFold,EvoscaleAI, Churchill Scholar@Cambridge_Uni & seen on Netflix
Posts
Media
Videos
Starter Packs
Pinned
Yousuf A. Khan
@yousufakhan.bsky.social
· Aug 27
Yousuf A. Khan (YAK) Lab - RNA Research at Stanford University
Yousuf A. Khan (YAK) Lab - Studying RNA folding, interactions, and cellular effects using Molecular and Cellular Physiology, Structural Biology, and Machine Learning at Stanford University
www.yousufakhan.com
Yousuf A. Khan
@yousufakhan.bsky.social
· Aug 27
Yousuf A. Khan
@yousufakhan.bsky.social
· Aug 27
Yousuf A. Khan (YAK) Lab - RNA Research at Stanford University
Yousuf A. Khan (YAK) Lab - Studying RNA folding, interactions, and cellular effects using Molecular and Cellular Physiology, Structural Biology, and Machine Learning at Stanford University
www.yousufakhan.com
Reposted by Yousuf A. Khan
Yousuf A. Khan
@yousufakhan.bsky.social
· Jul 29
Reposted by Yousuf A. Khan
Alex Rubinsteyn
@alexr.bsky.social
· Jul 23
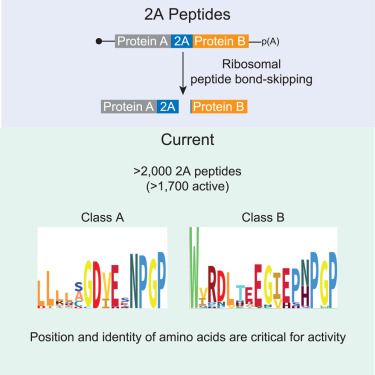
Systematic identification and characterization of eukaryotic and viral 2A peptide-bond-skipping sequences
Rao et al. identified thousands of previously unknown 2A peptides across both viruses
and eukaryotes using an HMMER analysis. The authors further identified a unique class
of 2A peptides, class B, who...
www.cell.com
Reposted by Yousuf A. Khan