Schapiro Lab
@schapirolab.bsky.social
340 followers
410 following
38 posts
Schapiro lab at Heidelberg University and Heidelberg University Hospital. We work on #HighlyMultiplexedImaging and #SpatialOmics
Posts
Media
Videos
Starter Packs
Pinned
Schapiro Lab
@schapirolab.bsky.social
· Dec 18
Reposted by Schapiro Lab
Schapiro Lab
@schapirolab.bsky.social
· Sep 13
Lukas Hatscher
@loggas.bsky.social
· Sep 13

Highly multiplexed imaging recovers immune and metabolic niches in multiple myeloma associated with disease progression and bone involvement
Multiple myeloma (MM) is a cancer arising from genetically aberrant plasma cells (PCs) that remain dependent on the bone marrow (BM) microenvironment for disease establishment and progression. Here, w...
www.biorxiv.org
Reposted by Schapiro Lab
Schapiro Lab
@schapirolab.bsky.social
· Aug 19
Internationaler Austausch als hervorragendes Beispiel für gemeinschaftliches Lernen - Universität Heidelberg
Benjamin Rombaut, Bioinformatik – Doktorand in der Forschungsgruppe „Data Mining and Modelling for Biomedicine“ am VIB-UGent Zentrum für Entzündungsforschung an
www.uni-heidelberg.de
Reposted by Schapiro Lab
Schapiro Lab
@schapirolab.bsky.social
· Mar 7

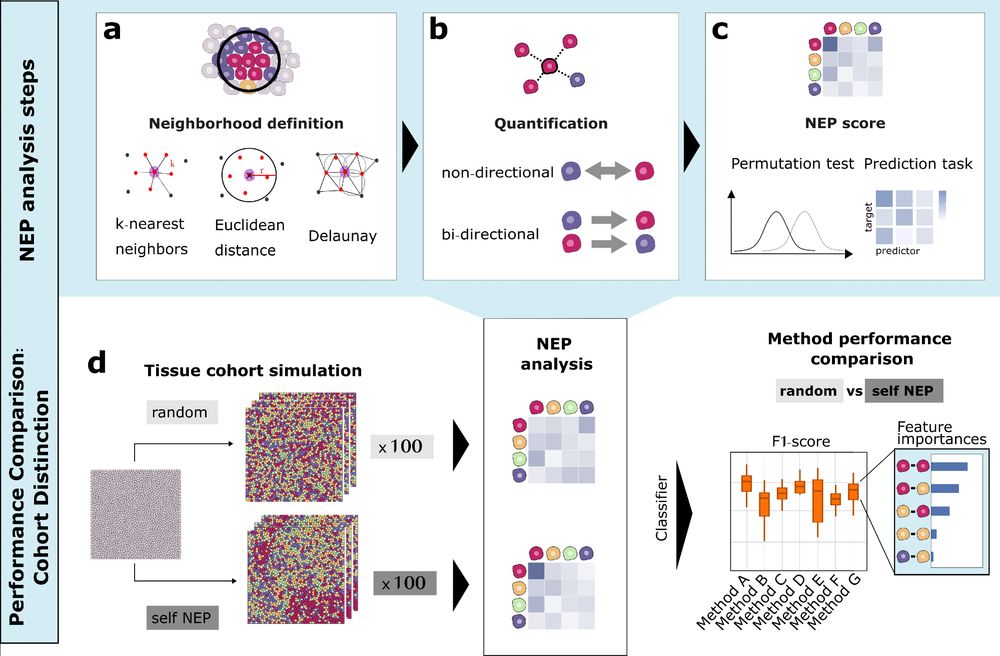
micronuclAI enables automated quantification of micronuclei for assessment of chromosomal instability - Communications Biology
micronuclAI, a deep-learning based pipeline, enables automated and reliable quantification of micronuclei from nuclei-stained human and murine cancer cell lines for quantification of chromosomal insta...
www.nature.com
Schapiro Lab
@schapirolab.bsky.social
· Mar 7