Samuel Gunz
@samuelgunz.bsky.social
130 followers
380 following
13 posts
PhD student in Statistical Bioinformatics at the University of Zurich and the Swiss Institue of Bioinformatics
Posts
Media
Videos
Starter Packs
Reposted by Samuel Gunz
Martin Emons
@martinemons.bsky.social
· Sep 11

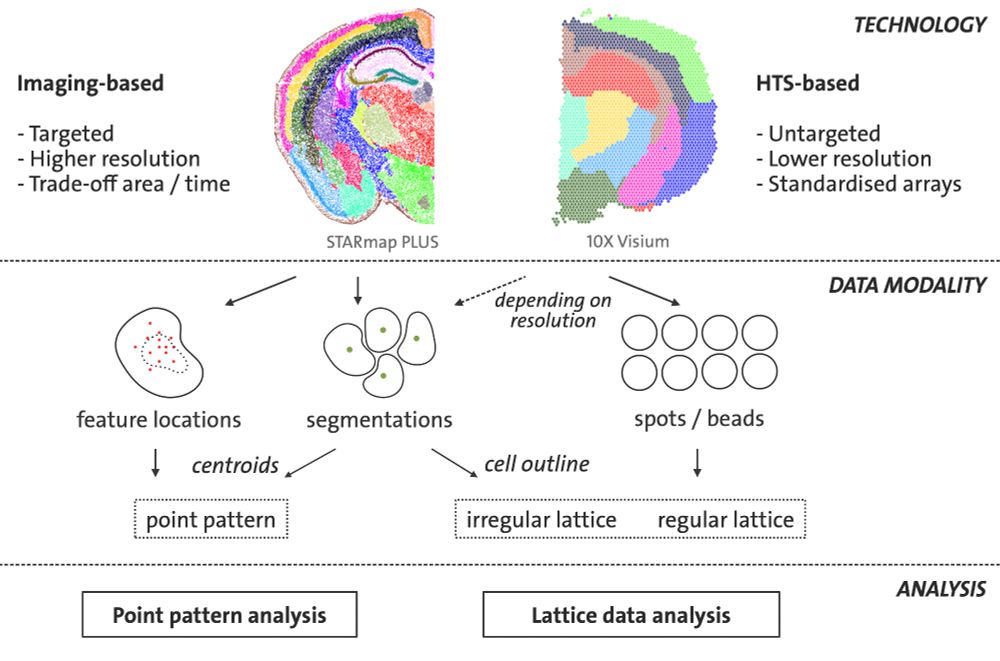
Harnessing the potential of spatial statistics for spatial omics data with pasta
Abstract. Spatial omics allow for the molecular characterization of cells in their spatial context. Notably, the two main technological streams, imaging-ba
academic.oup.com
Reposted by Samuel Gunz
Martin Emons
@martinemons.bsky.social
· Jun 27

Harnessing the Potential of Spatial Statistics for Spatial Omics Data with pasta
Spatial omics assays allow for the molecular characterisation of cells in their spatial context. Notably, the two main technological streams, imaging-based and high-throughput sequencing-based, can gi...
arxiv.org
Samuel Gunz
@samuelgunz.bsky.social
· Dec 4
Samuel Gunz
@samuelgunz.bsky.social
· Dec 4
Samuel Gunz
@samuelgunz.bsky.social
· Dec 4
Samuel Gunz
@samuelgunz.bsky.social
· Dec 4
Samuel Gunz
@samuelgunz.bsky.social
· Dec 4
Samuel Gunz
@samuelgunz.bsky.social
· Dec 4

pasta: Pattern Analysis for Spatial Omics Data
Spatial omics assays allow for the molecular characterisation of cells in their spatial context. Notably, the two main technological streams, imaging-based and high-throughput sequencing-based, can gi...
arxiv.org