Nezar Abdennur
@nvictus.bsky.social
100 followers
150 following
24 posts
computational biologist / biological computer / asst prof @UMassChan / phd @MIT / http://abdenlab.org
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Nezar Abdennur
Reposted by Nezar Abdennur
Nezar Abdennur
@nvictus.bsky.social
· Aug 11
Nezar Abdennur
@nvictus.bsky.social
· Aug 11
Nezar Abdennur
@nvictus.bsky.social
· Aug 11

GitHub - abdenlab/jointly-hic: Genomics research toolkit for jointly embedding Hi-C 3D chromatin contact matrices into the same vector space
Genomics research toolkit for jointly embedding Hi-C 3D chromatin contact matrices into the same vector space - abdenlab/jointly-hic
github.com
Nezar Abdennur
@nvictus.bsky.social
· Aug 11

Joint decomposition of Hi-C maps reveals salient features of genome architecture across tissues and development
The spatial organization of chromosomes in the nucleus is fundamental to cellular processes. Contact frequency maps from Hi-C and related chromosome conformation capture assays are increasingly availa...
www.biorxiv.org
Reposted by Nezar Abdennur
Reposted by Nezar Abdennur
Nezar Abdennur
@nvictus.bsky.social
· Jul 7
Nezar Abdennur
@nvictus.bsky.social
· Jul 7
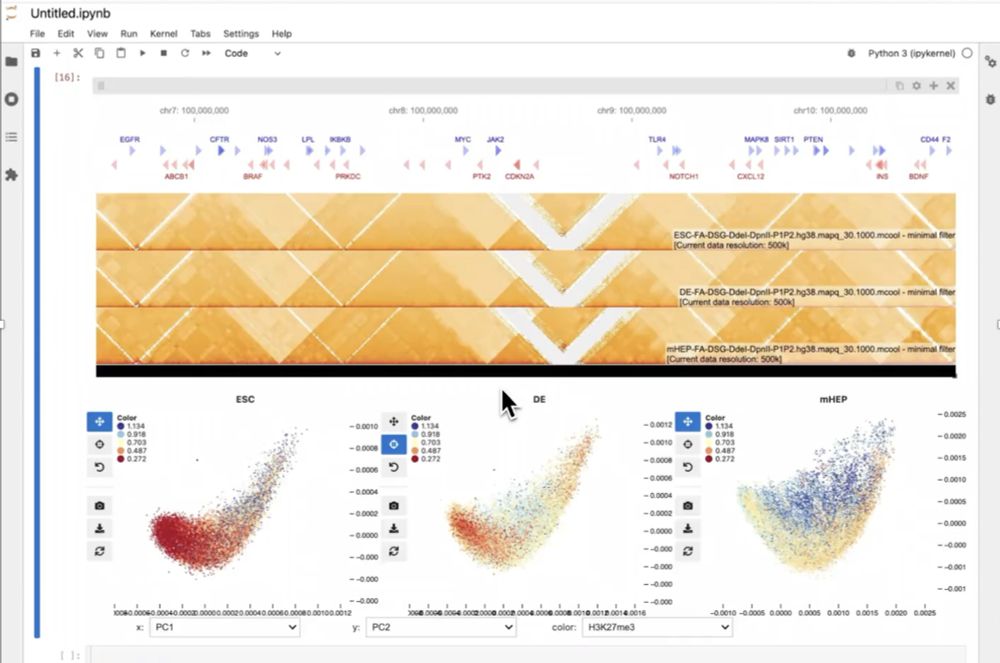
Breaking the silo: composable bioinformatics through cross-disciplinary open standards SciPy 2025
The practice of data science in genomics and computational biology is fraught with friction. This is in large part because bioinformatic tools tend to be tightly coupled to file input/output. As a res...
cfp.scipy.org
Nezar Abdennur
@nvictus.bsky.social
· Jul 7
Nezar Abdennur
@nvictus.bsky.social
· Jul 7
Nezar Abdennur
@nvictus.bsky.social
· Jul 7
Nezar Abdennur
@nvictus.bsky.social
· Jul 7
Nezar Abdennur
@nvictus.bsky.social
· Jul 7

Breaking the silo: composable bioinformatics through cross-disciplinary open standards SciPy 2025
The practice of data science in genomics and computational biology is fraught with friction. This is in large part because bioinformatic tools tend to be tightly coupled to file input/output. As a res...
cfp.scipy.org
Nezar Abdennur
@nvictus.bsky.social
· Jul 7
Nezar Abdennur
@nvictus.bsky.social
· Jul 7
Nezar Abdennur
@nvictus.bsky.social
· Jul 7