Janani Durairaj (Jay)
@ninjani.bsky.social
1.2K followers
380 following
32 posts
Computational biologist @biozentrum.bsky.social. Likes protein structures.
https://ninjani.github.io/
Posts
Media
Videos
Starter Packs
Pinned
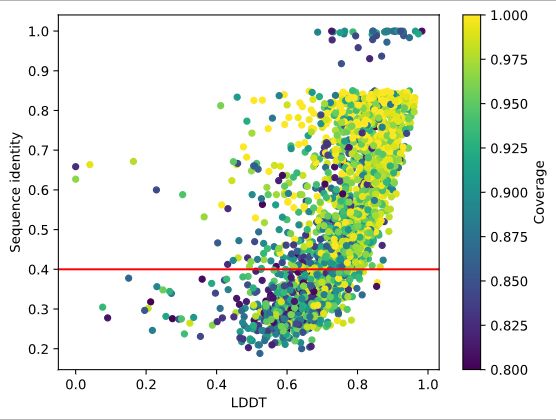
Excited to share our latest preprint evaluating AlphaFold3, Boltz-1, Chai-1 and Protenix for predicting protein-ligand interactions, featuring our newly introduced benchmark dataset 🌹Runs N’ Poses🌹!
www.biorxiv.org/content/10.1...
🧵👇 (1/n)
www.biorxiv.org/content/10.1...
🧵👇 (1/n)

Have protein-ligand co-folding methods moved beyond memorisation?
Deep learning has driven major breakthroughs in protein structure prediction, however the next critical advance is accurately predicting how proteins interact with other molecules, especially small mo...
www.biorxiv.org
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Roni Odai
@rodai.bsky.social
· Oct 2

The Viral AlphaFold Database of monomers and homodimers reveals conserved protein folds in viruses of bacteria, archaea, and eukaryotes
VAD is a Viral AlphaFold Database of protein monomers and homodimers from viruses infecting hosts across the tree of life.
www.science.org
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
🔬 Workshop: Future of Structure Prediction Benchmarking
📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.
📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.

The future of structure prediction benchmarking: measuring progress and breakthroughs · Luma
Benchmarking has been a key driver of progress in protein structure prediction methods. As the field continues to evolve, several key questions prevail:
How…
lu.ma
Reposted by Janani Durairaj (Jay)
🔬 Workshop: Future of Structure Prediction Benchmarking
📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.
📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.

The future of structure prediction benchmarking: measuring progress and breakthroughs · Luma
Benchmarking has been a key driver of progress in protein structure prediction methods. As the field continues to evolve, several key questions prevail:
How…
lu.ma
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Yo Akiyama
@yoakiyama.bsky.social
· Aug 5

Scaling down protein language modeling with MSA Pairformer
Recent efforts in protein language modeling have focused on scaling single-sequence models and their training data, requiring vast compute resources that limit accessibility. Although models that use ...
biorxiv.org
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Reposted by Janani Durairaj (Jay)
Introducing CAMEO Structures & Complexes - automated weekly blind benchmarking of structure prediction servers. Now with heteromeric and protein-ligand complexes.
Join us and register your server now!
cameo3d.org
Join us and register your server now!
cameo3d.org
Reposted by Janani Durairaj (Jay)
The Viral AlphaFold Database of monomers and homodimers reveals conserved protein folds in viruses of bacteria, archaea, and eukaryotes https://www.biorxiv.org/content/10.1101/2025.05.14.653371v1
Reposted by Janani Durairaj (Jay)
Arne Elofsson
@bioinfo.se
· May 5
Limits of deep-learning-based RNA prediction methods
Motivation: In recent years, tremendous advances have been made in predicting protein structures and protein-protein interactions. However, progress in predicting the structure of RNA, either alone or...
www.biorxiv.org
Introducing CAMEO Structures & Complexes - automated weekly blind benchmarking of structure prediction servers. Now with heteromeric and protein-ligand complexes.
Join us and register your server now!
cameo3d.org
Join us and register your server now!
cameo3d.org
Reposted by Janani Durairaj (Jay)