Antoine de Chevigny
@antoinedechevigny.bsky.social
120 followers
170 following
22 posts
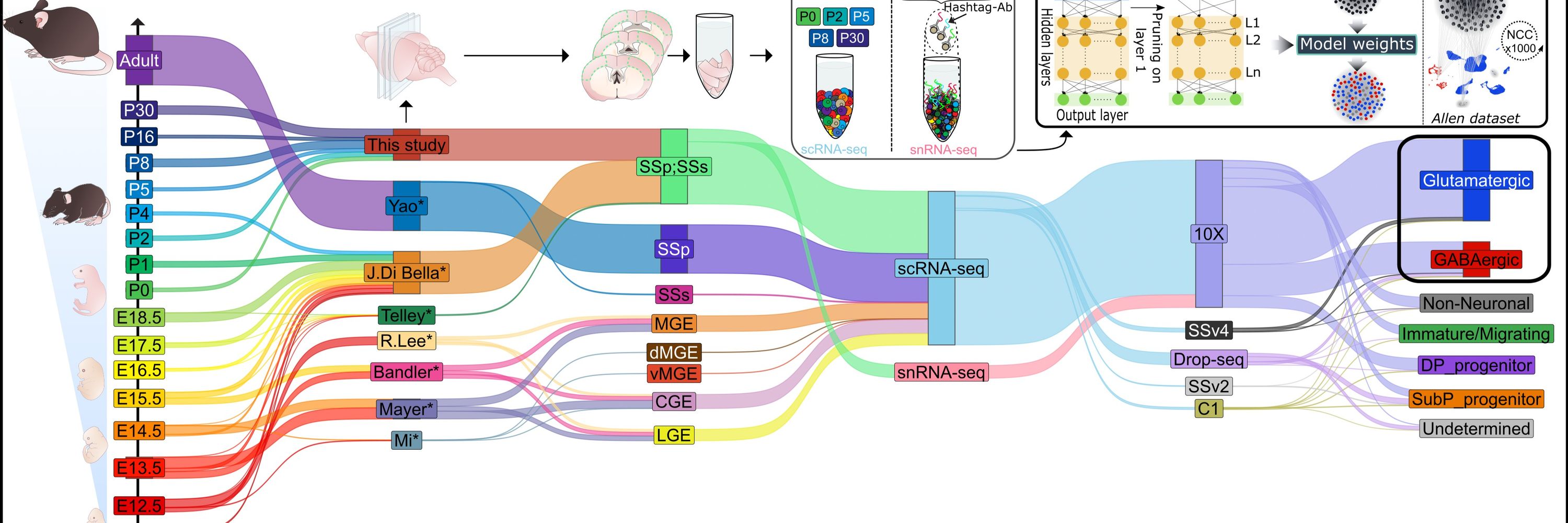
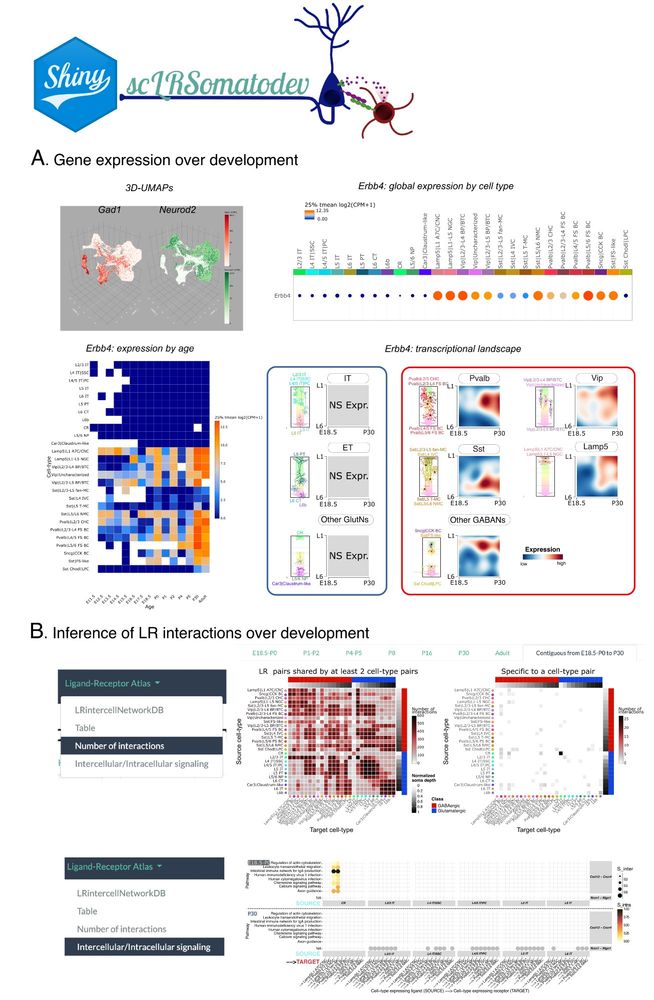
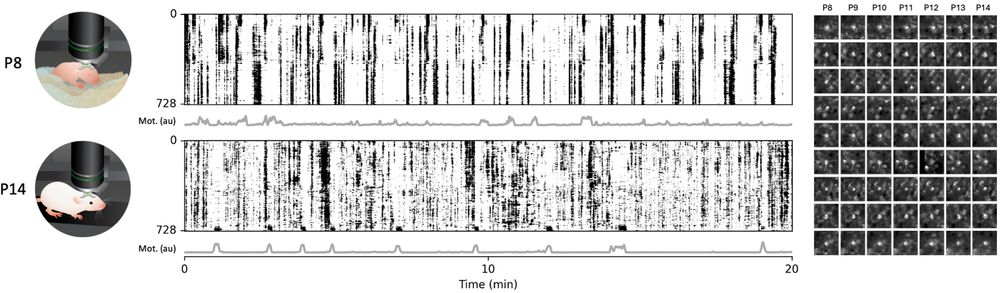
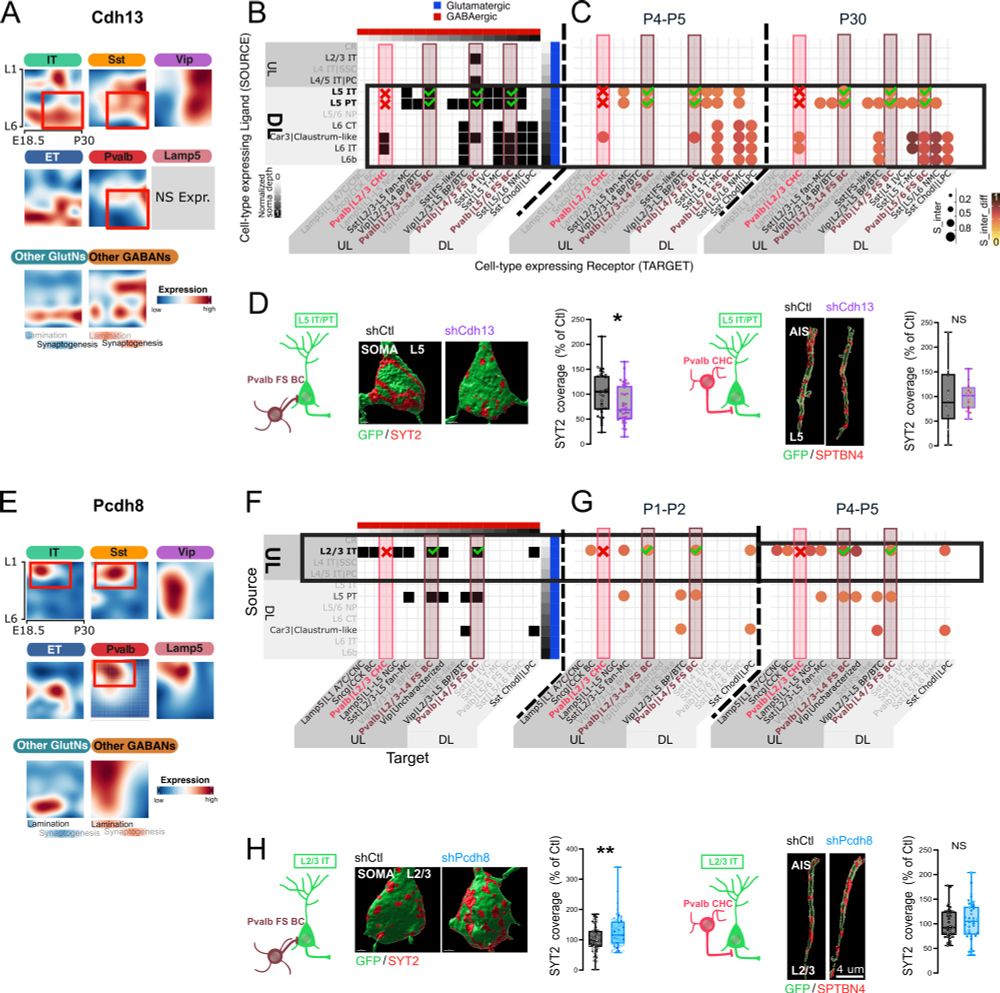
Development and function of the mammalian neocortex
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Antoine de Chevigny
Reposted by Antoine de Chevigny
Reposted by Antoine de Chevigny
Reposted by Antoine de Chevigny
Christophe 🔬 L
@christlet.bsky.social
· Jan 9

The Institute of Neurophysiopathology (INP) welcomes tenured researchers/lecturers, and junior researchers seeking a permanent position in France to lead or strengthen its research programs
The Institute of Neurophysiopathology (INP) welcomes tenured researchers/lecturers, and junior researchers seeking a permanent position in France to lead or strengthen its research programs RESEARCHER...
euraxess.ec.europa.eu
Reposted by Antoine de Chevigny
Cedric Boeckx
@cedricboeckx.bsky.social
· Feb 28
Reposted by Antoine de Chevigny
Riccardo Bocchi
@bocchiric.bsky.social
· Feb 24
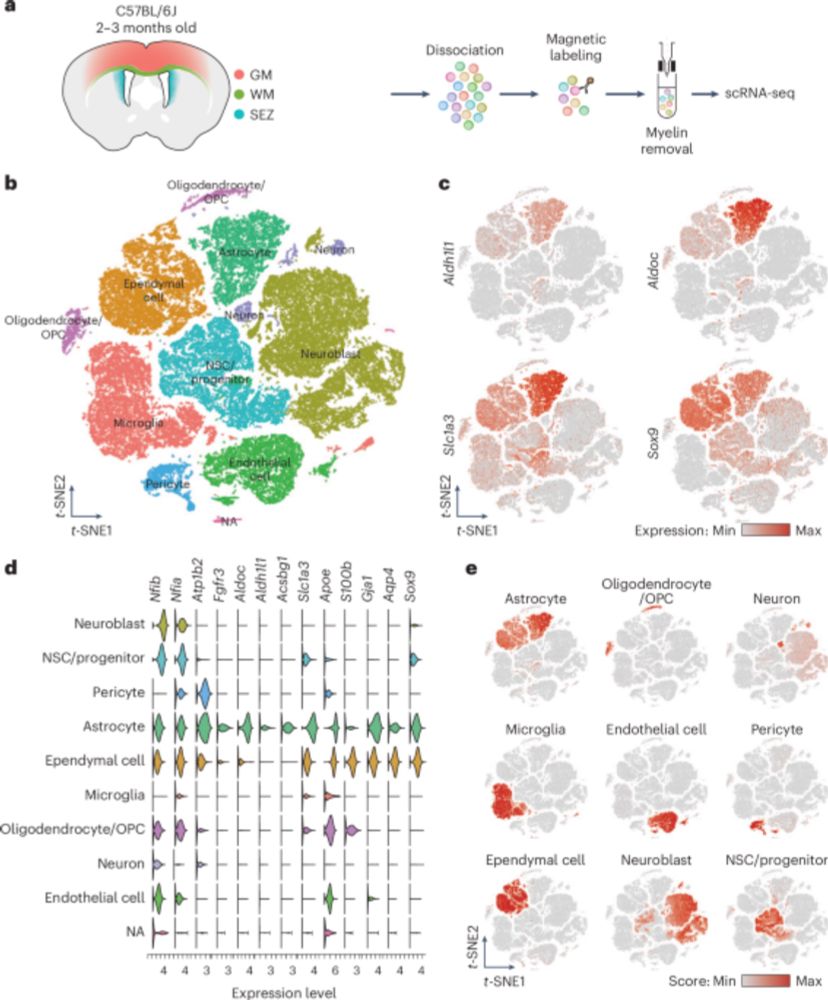
Astrocyte heterogeneity reveals region-specific astrogenesis in the white matter - Nature Neuroscience
White matter (WM) astrocytes differ significantly from gray matter astrocytes, with WM astrocytes in the forebrain exhibiting unique proliferation capacity, which is absent in cerebellar WM, suggestin...
www.nature.com
Reposted by Antoine de Chevigny
Reposted by Antoine de Chevigny
Reposted by Antoine de Chevigny
Eli Merriam
@elimerriam.bsky.social
· Feb 27
Brain-wide presynaptic networks of functionally distinct cortical neurons - Nature
Behavioural-state-dependent pyramidal neurons have a distinct pattern of long-range glutamatergic inputs, with a larger proportion of thalamic versus motor cortex inputs compared with non-behavio...
www.nature.com