Dominik Handler
@86dominik.bsky.social
250 followers
360 following
19 posts
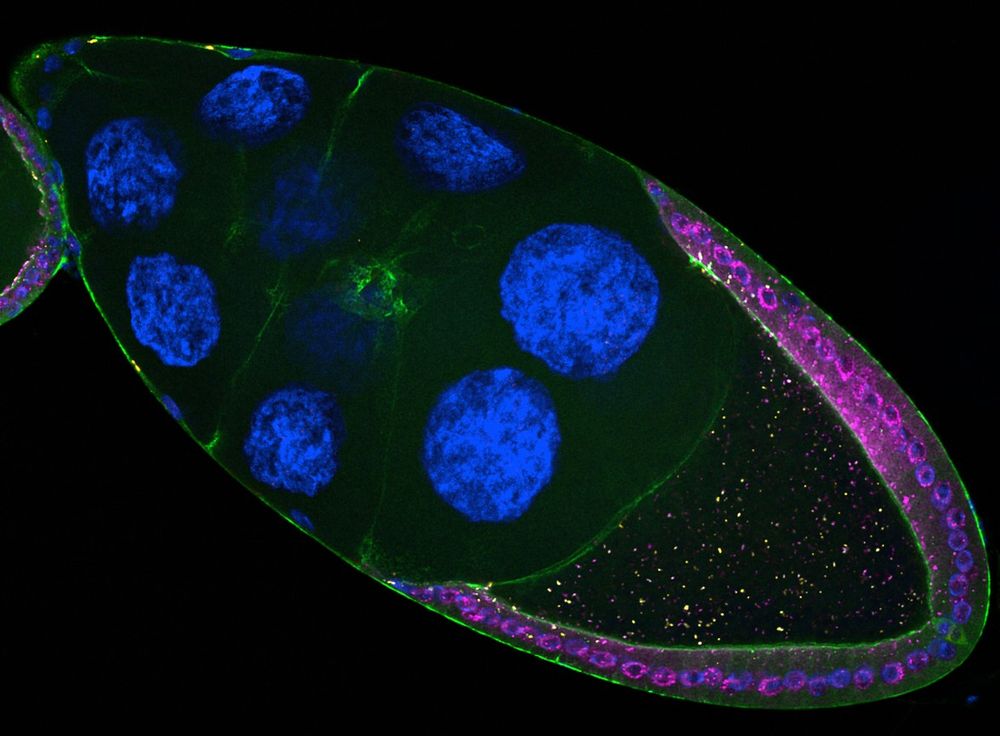
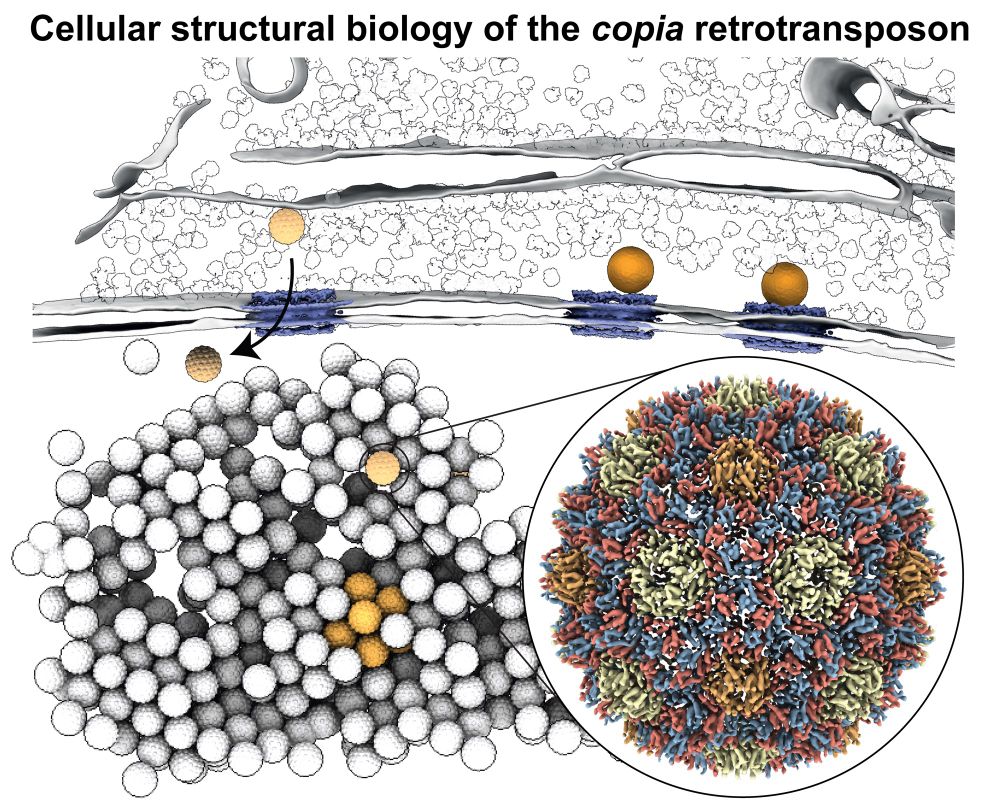
Drosophila genetic conflicts
piRNAs | transposons | genomics
Staff scientist in the Brennecke lab - Vienna
Posts
Media
Videos
Starter Packs
Reposted by Dominik Handler
Reposted by Dominik Handler
Dominik Handler
@86dominik.bsky.social
· Aug 21
Reposted by Dominik Handler
Reposted by Dominik Handler
Reposted by Dominik Handler
Reposted by Dominik Handler
Reposted by Dominik Handler
Reposted by Dominik Handler